UANanoDock: A Web-Based UnitedAtom Multiscale Nanodocking Tool for Predicting Protein Adsorption onto Nanoparticles
3 April 2025Abstract
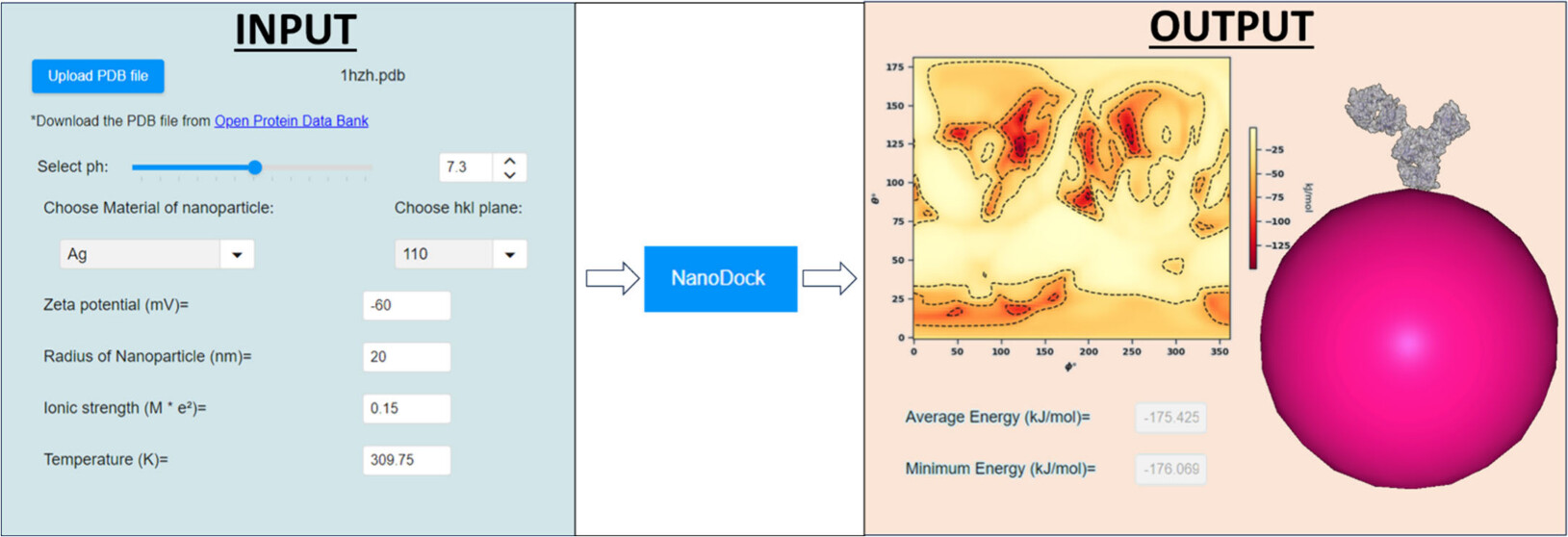
UANanoDock is a web-based application with a graphical user interface designed for modeling protein–nanomaterial interactions, accessible via the Enalos Cloud Platform (https://www.enaloscloud.novamechanics.com/compsafenano/uananodock/). The application’s foundation lies in the UnitedAtom multiscale model, previously reported for predicting the adsorption energies of biopolymers and small molecules onto nanoparticles (NPs). UANanoDock offers insights into optimal protein orientations when bound to spherical NP surfaces, considering factors such as material type, NP radius, surface potential, and amino acid (AA) ionization states at specific pH levels. The tool’s computational time is determined solely by the protein’s AA count, regardless of NP size. With its efficiency (e.g., approximately 60 s processing time for a 1331 AA protein) and versatility (accommodating any protein with a standard AA sequence in PDB format), UANanoDock serves as a prescreening tool for identifying proteins likely to adsorb onto NP surfaces. An illustration of UANanoDock’s utility is provided, demonstrating its application in the rational design of immunoassays by determining the preferred orientation of the immunoglobulin G (IgG) antibody adsorbed on Ag NPs.
Reference
Subbotina, J., Kolokathis, P.D., Tsoumanis, A., Sidiropoulos, N.K., Rouse, I., Lynch, I., Lobaskin, V., & Afantitis, A. (2025). UANanoDock: A Web-Based UnitedAtom Multiscale Nanodocking Tool for Predicting Protein Adsorption onto Nanoparticles. Journal of Chemical Information and Modeling. https://doi.org/10.1021/acs.jcim.4c02292